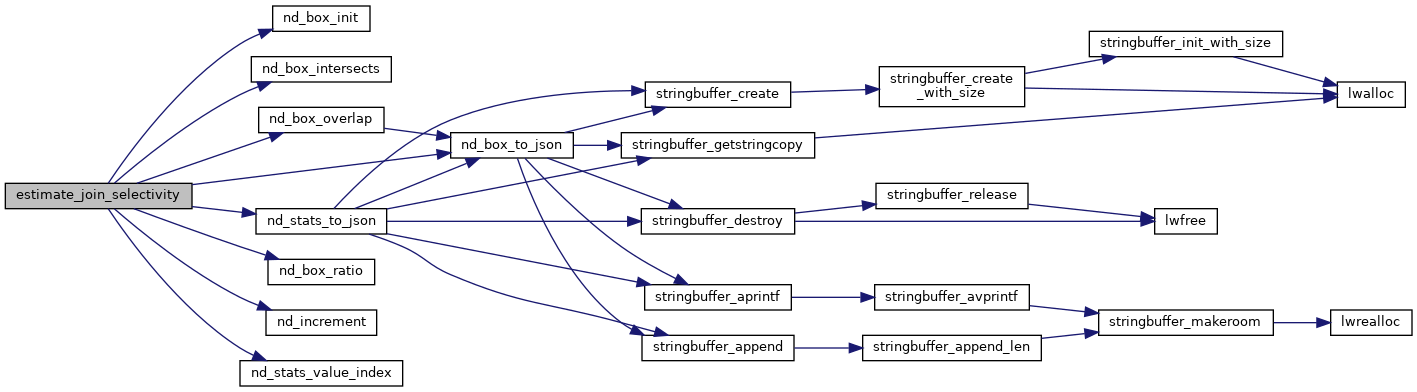
Given two statistics histograms, what is the selectivity of a join driven by the && or &&& operator?
Join selectivity is defined as the number of rows returned by the join operator divided by the number of rows that an unconstrained join would return (nrows1*nrows2).
To get the estimate of join rows, we walk through the cells of one histogram, and multiply the cell value by the proportion of the cells in the other histogram the cell overlaps: val += val1 * ( val2 * overlap_ratio )
891{
892 int ncells1, ncells2;
893 int ndims1, ndims2, ndims;
894 double ntuples_max;
895 double ntuples_not_null1, ntuples_not_null2;
896
909 int d;
910 double val = 0;
911 float8 selectivity;
912
913
914 if ( ! ( s1 && s2 ) )
915 {
916 elog(NOTICE, " estimate_join_selectivity called with null inputs");
918 }
919
920
923
924
925 if ( ncells1 > ncells2 )
926 {
928 s1 = s2;
929 s2 = stats_tmp;
930 }
931
934
935
938
939
940
943 ntuples_max = ntuples_not_null1 * ntuples_not_null2;
944
945
946 ndims1 = (int)roundf(s1->
ndims);
947 ndims2 = (int)roundf(s2->
ndims);
948 ndims = Max(ndims1, ndims2);
949
950
953
954
956 {
957 POSTGIS_DEBUG(3, "relation stats do not intersect, returning 0");
958 PG_RETURN_FLOAT8(0.0);
959 }
960
961
962
963
964
966 {
967 POSTGIS_DEBUG(3, "could not calculate overlap of relations");
969 }
970
971
972 for ( d = 0; d < ndims1; d++ )
973 {
974 at1[d] = ibox1.
min[d];
977 size1[d] = (int)roundf(s1->
size[d]);
978 cellsize1[d] = width1[d] / size1[d];
979 }
980
981
982 for ( d = 0; d < ndims2; d++ )
983 {
986 size2[d] = (int)roundf(s2->
size[d]);
987 cellsize2[d] = width2[d] / size2[d];
988 }
989
990
991 do
992 {
993 double val1;
994
997 for ( d = 0; d < ndims1; d++ )
998 {
999 nd_cell1.
min[d] = min1[d] + (at1[d]+0) * cellsize1[d];
1000 nd_cell1.
max[d] = min1[d] + (at1[d]+1) * cellsize1[d];
1001 }
1002
1003
1005
1006
1007 for ( d = 0; d < ndims2; d++ )
1008 {
1009 at2[d] = ibox2.
min[d];
1010 }
1011
1012 POSTGIS_DEBUGF(3,
"at1 %d,%d %s", at1[0], at1[1],
nd_box_to_json(&nd_cell1, ndims1));
1013
1014
1016
1017
1018 do
1019 {
1020 double ratio2;
1021 double val2;
1022
1023
1026 for ( d = 0; d < ndims2; d++ )
1027 {
1028 nd_cell2.
min[d] = min2[d] + (at2[d]+0) * cellsize2[d];
1029 nd_cell2.
max[d] = min2[d] + (at2[d]+1) * cellsize2[d];
1030 }
1031
1032 POSTGIS_DEBUGF(3,
" at2 %d,%d %s", at2[0], at2[1],
nd_box_to_json(&nd_cell2, ndims2));
1033
1034
1035 ratio2 =
nd_box_ratio(&nd_cell1, &nd_cell2, Max(ndims1, ndims2));
1036
1037
1039 POSTGIS_DEBUGF(3, " val1 %.6g val2 %.6g ratio %.6g", val1, val2, ratio2);
1040 val += val1 * (val2 * ratio2);
1041 }
1043
1044 }
1046
1047 POSTGIS_DEBUGF(3, "val of histogram = %g", val);
1048
1049
1050
1051
1052
1053
1054
1057
1058 POSTGIS_DEBUGF(3, "val scaled to full table size = %g", val);
1059
1060
1061
1062
1063
1064
1065
1066
1067
1068
1069
1070
1071
1072
1073
1074
1075 selectivity = val / ntuples_max;
1076
1077
1078 if ( isnan(selectivity) || ! isfinite(selectivity) || selectivity < 0.0 )
1079 {
1081 }
1082 else if ( selectivity > 1.0 )
1083 {
1084 selectivity = 1.0;
1085 }
1086
1087 return selectivity;
1088}
static int nd_box_intersects(const ND_BOX *a, const ND_BOX *b, int ndims)
Return true if ND_BOX a overlaps b, false otherwise.
static int nd_increment(ND_IBOX *ibox, int ndims, int *counter)
Given an n-d index array (counter), and a domain to increment it in (ibox) increment it by one,...
static char * nd_box_to_json(const ND_BOX *nd_box, int ndims)
Convert an ND_BOX to a JSON string for printing.
static char * nd_stats_to_json(const ND_STATS *nd_stats)
Convert an ND_STATS to a JSON representation for external use.
#define DEFAULT_ND_JOINSEL
#define FALLBACK_ND_SEL
More modest fallback selectivity factor.
#define FALLBACK_ND_JOINSEL
static int nd_box_init(ND_BOX *a)
Zero out an ND_BOX.
static int nd_box_overlap(const ND_STATS *nd_stats, const ND_BOX *nd_box, ND_IBOX *nd_ibox)
What stats cells overlap with this ND_BOX? Put the lowest cell addresses in ND_IBOX->min and the high...
static double nd_box_ratio(const ND_BOX *cover, const ND_BOX *target, int ndims)
static int nd_stats_value_index(const ND_STATS *stats, const int *indexes)